Project Description
Diffusion MRI (dMRI) has the ability to detect white matter abnormalities in-vivo by quantifying the water diffusivity that is directionally restricted by barriers such as axonal membranes, myelin and cell packing. In dMRI, the challenge is to describe the 3D probability density function (often referred to as the diffusion propagator) of the displacement of water molecules in a restricted environment. The dMRI signal and the diffusion propagator are related by the 3D Fourier transform. So, one approach is to directly sample the entire Fourier do- main (q-space) and then numerically compute the diffusion propagator. q-space can also be sampled on a sphere of radius |q| (a shell), enabling measurement of the angular distribution of the diffusion propagator. By sampling on several spherical shells with different radii, one can recover the radial part of the diffusion propagator, which encodes the type of diffusion (hindered vs. restricted), allowing estimation of new microstructural biomarkers.
The standard approach to studying the neuroanatomy of pediatric brains has been using DTI, which allows scanning a subject in about 2 minutes with 6-10 gradient directions. DTI describes the diffusion propagator with six parameters, but the information provided by these parameters is limited and non-specific. Recently, diffusion models that go beyond DTI were proposed to extract important biomarkers such as compartmentalization and zero displacement probability, however, there are technical challenges that must be overcome to move beyond DTI in pediatric clinical research.
In particular, dMRI of pediatric patients presents several challenges; chief among them is the inability of children to stay still in an MRI scanner for more than 5 minutes. The problem is further exacerbated if the children are hyperactive as in ADHD. This is a critical barrier for advanced dMRI acquisitions where motion cannot be tolerated for the typical 10-15 minute scan time. Further, if one wants to obtain information about the radial part of the diffusion propagator, a dense sampling of the q-space with high b-values is required, which results in even longer acquisition times (40-50 minutes). One alternative is to use sedation and anesthesia on children (or other types of patients with dementia) who undergo this long scan protocol. However, as has been well documented in several studies, sedation can fail in 30-40% of the cases, while around 20% of children experience an adverse event. Hence, sedation is not a particularly safe option for use during dMRI acquisition.
Thus, there is a critical need to reduce the acquisition time so that detailed analysis of the brain microstructure is possible to study any neurological disease in the pediatric population.
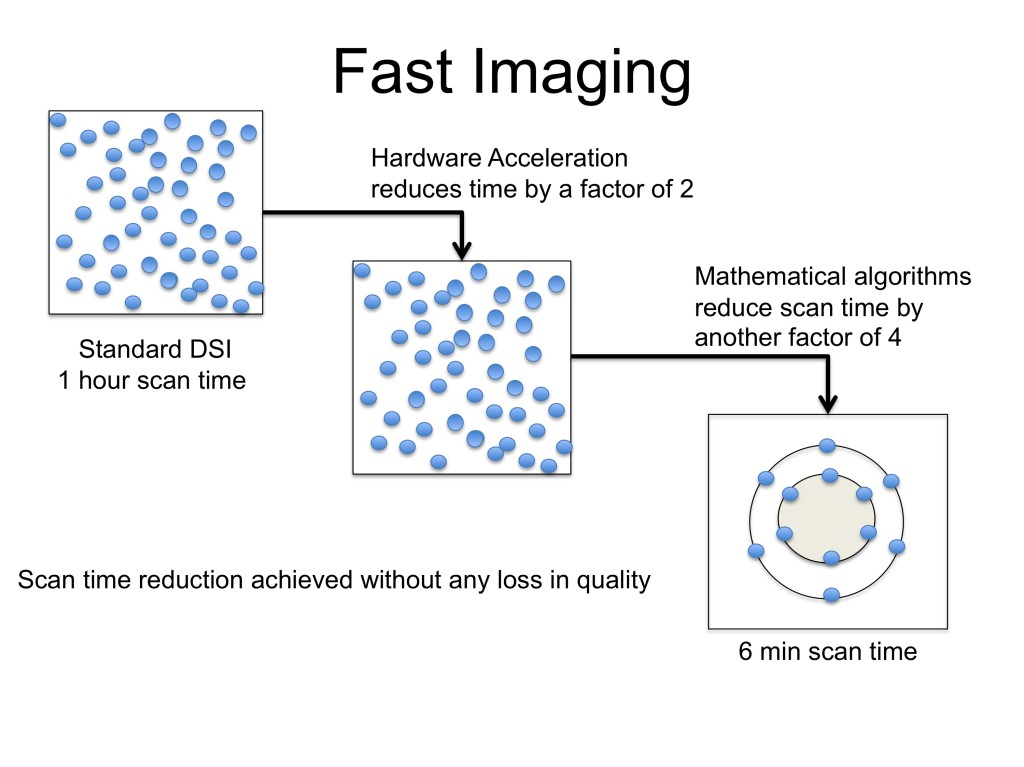 We address the above limitations of long scan time for single shell (HARDI) and multi-shell diffusion imaging (MSDI) protocols using a two pronged strategy. By utilizing the concepts of compressive sampling (CS) to sparsely sample the q-space and combining it with a multi-slice acquisition protocol, we reduce the number of measurements while acquiring them simultaneously. This results in significant reduction in scan time for HARDI and MSDI protocols. HARDI acquisition of pediatric patients will enable analysis of complex fiber pathways, while MSDI acquisition will reveal compartmental information as well as other markers of tissue structure which have not been measured before in clinical research. These measures have been shown to be more sensitive to the underlying anatomy than the standard DTI measures.
We address the above limitations of long scan time for single shell (HARDI) and multi-shell diffusion imaging (MSDI) protocols using a two pronged strategy. By utilizing the concepts of compressive sampling (CS) to sparsely sample the q-space and combining it with a multi-slice acquisition protocol, we reduce the number of measurements while acquiring them simultaneously. This results in significant reduction in scan time for HARDI and MSDI protocols. HARDI acquisition of pediatric patients will enable analysis of complex fiber pathways, while MSDI acquisition will reveal compartmental information as well as other markers of tissue structure which have not been measured before in clinical research. These measures have been shown to be more sensitive to the underlying anatomy than the standard DTI measures.
Decreased scan time provides two principal advantages: currently possible scans are faster, and currently impossible protocols (due to clinically infeasible scan times) may be attempted. We have quantitatively shown that using compressive sampling, the total number of samples over the different shells (for MSDI) are similar to today’s single-shell acquisitions, thereby facilitating its translation to the clinic and replacing existing protocols with a more informative one.
In order to validate the “compressed sensing” approach, we organized a SPARC (Sparse Reconstuction Challenge) dMRI to test the performance of various methods on data acquired from a physical phantom, with different levels of “compression” or “sparsity”. As seen from the results, the methods developed at the PNL (in collaboration with LMI) performed better than any other approach that participated in the challenge. More details about the challenge are given at:
http://projects.iq.harvard.edu/sparcdmri/results
Technical details about our method are given in this presentation.






Comments are closed.